|
Phylogenetic Disease Ecology of Plants Ingrid M. Parker & Gregory S. Gilbert
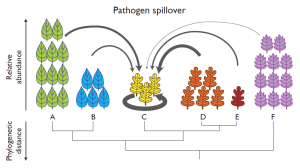
Pathogens can often attack multiple species. This could affect how pathogens spread and the impact of the diseases they cause. Two host species are more likely to share a pathogen when they are closely related to each other. This research explores three important predictions that follow from this simple idea. The first is that in nature, the complex networks of plants and their pathogens will be structured in predictable ways that reflect the evolutionary relationships among the species. Second, when a new plant species (such as a crop) is introduced into an area, pathogens will “spill over” from close relatives already growing there. Third, because pathogens are less likely to spread between species that are not closely related, mixtures of crop species could be designed to suppress disease pressure in agriculture. In the process of testing these predictions, the we will develop new analytical tools for the study of plant disease and for the protection of agriculture and other plant resources. The large training component of this project will include high school classes, college classes, senior thesis students, and PhD students. Participating students will engage in active, experiential learning in forest ecology, ecological monitoring, agroecology and ecological horticulture, and both laboratory and bioinformatics aspects of metagenomics. For each of the three components of the project, we will take advantage of a quantitative model previously developed and validated that predicts how pathogen host ranges are constrained by the evolutionary relationships among host plants. In a novel application of network theory, we will incorporate phylogenetic structure into network models for plant-pathogen interactions. We will use a metagenomics approach (identifying all fungal species growing inside plants by extracting and sequencing their DNA, sometimes called an eDNA approach) to characterize the plant-pathogen networks of three plant communities and will test predictions against the empirical networks. Novel hosts will be used in a transplant experiment in the field and an inoculation experiment in the laboratory to measure which local fungi colonize novel hosts, and how the phylogenetic structure of local plant-fungus networks affects fungal spillover to novel hosts. Finally, collaborating with the UCSC CASFS certified organic research farm, we will test whether the “Dilution Effect” lowers disease pressure in cover crop mixtures, and whether greater phylogenetic distance among intercropped species suppresses disease. |
|
|
|
Stephen Hubbell, Brant Faircloth, Gregory S. Gilbert, and Travis Glenn |
|
This project integrates taxonomic, genetic, and functional approaches in a combination of observations and experiments to test the Enemy Susceptibility Hypothesis (ESH) for tropical tree diversity and rarity. This hypothesis predicts that rare tree species will have a greater percentage of hollow-trunk trees, harbor more pathogenic fungal species, and also share more of these pathogens with other tree species, than common tree species. At forest plots on Barro Colorado Island, Panama, we used sonic tomography to examine thousands of living trees, detect internal decay, and sample fungi from wood and leaves to determine which fungal species are present on which trees. We will be experimentally testing pathogenicity, as well as looking at expression of lignin-degrading enzymes in the fungi, and how they may be involved in host ranges. If the Enemy Susceptibility Hypothesis is true, it may provide a general explanation for the high diversity and rarity of tree species in tropical forests worldwide. The project also includes experiential, inquiry-based learning for a number of Panamanian interns. The project has also incorporated two 10-day field courses for high school students, college students, and high school science teachers from California, Georgia, and Panama. The participants conducted research in the rain forest for 10 days, learning about tropical ecology, tree diseases, and molecular approaches to field ecology. |
|
|
And here is a recent talk that explores the connections between the Fungal Dimensions and Rare-species Advantage work.You Tube: Talk at Estación Biólogica de Doñana, Sevilla, Spain 20Jan2016